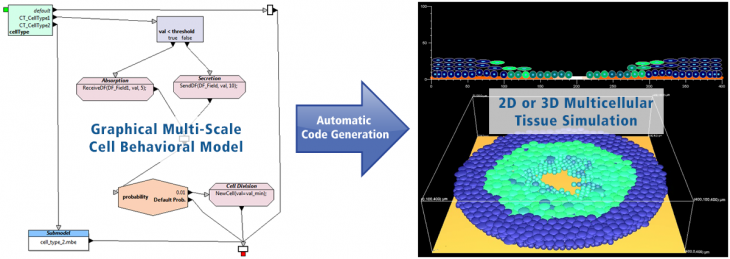
The EPISIM Platform provides the two ready-to-use and easy-to-install software systems EPISIM Modeller and EPISIM Simulator. EPISIM Modeller enables graphical multi-scale modeling of cellular behaviour in a multicellular context using process diagrams. The graphical cell behavioral models are automatically translated into executable code. This code can be loaded with EPISIM Simulator to conduct either a 2D or a 3D tissue simulation.
Cellular behavior is modeled with process diagrams using deterministic and / or stochastic model elements. SBML based quantiative subcellular models can be automatically imported and semantically integrated in a cell behavioral model. The EPISIM platform enables users to build their own cell behavioral model and tissue simulation in a purely graphical way. No dedicated programming skills are needed.
How to cite EPISIM:Sütterlin T, Kolb C, Dickhaus H, Jäger D, Grabe N (2013). Bridging the scales: semantic integration of quantitative SBML in graphical multi-cellular models and simulations with EPISIM and COPASI. Bioinformatics. 2013 Jan 15;29(2):223-9. Epub 2012 Nov 18. PubMed PMID: 23162085. |
EPISIM Movies
| EPISIM Simulation Demo Movies |
EPISIM Introductory Video Tutorials |
EPISIM Semantic SBML Model Integration Tutorials |
 |
 |
 |
Selected EPISIM Publications
Sütterlin T, Tsingos E, Bensaci J, Stamatas GN, Grabe N (2017). A 3D self-organizing
multicellular epidermis model of barrier formation and hydration with realistic
cell morphology based on EPISIM. Sci Rep. 2017 Mar 6;7:43472. doi:
10.1038/srep43472.
PubMed PMID: 28262741; PubMed Central PMCID: PMC5338006.
electronic version
Sütterlin T and Grabe N (2014). Graphical Multi-Scale Modeling of Epidermal Homeostasis with EPISIM. In, Querleux,B. (Ed.), Computational Biophysics of the Skin. Pan Stanford Publishing, Singapore, p. 421–460.
Sütterlin T,
Kolb C, Dickhaus H, Jäger D, Grabe N (2013). Bridging the scales:
semantic integration of quantitative SBML in graphical multi-cellular
models and simulations with EPISIM and COPASI. Bioinformatics. 2013 Jan
15;29(2):223-9. Epub 2012 Nov 18. PubMed
PMID: 23162085.
electronic version supplements
Safferling K,
Sütterlin T, Westphal K, Ernst C, Breuhahn K, James M, Jäger D, Halama N und Grabe
N (2013). Wound healing
revised: A novel reepithelialization mechanism revealed by in vitro and in
silico models. Journal of Cell Biology, 203 (4), pp. 691–709.
electronic version
Sütterlin T, Huber S,
Dickhaus H and Grabe N. (2009) Modeling multi-cellular behavior in
epidermal tissue homeostasis via finite state machines in multi-agent
systems, Bioinformatics, 25, 2057-2063.
PMID: 19535533
electronic version supplements
Acknowlegements
EPISIM would not exist if there were no freely available software libraries and frameworks. EPISIM is based on the following software being maintained and developed at university institutions:
 |
 |
 |
Moreover, EPISIM utilizes amongst others the following freely available platforms and libraries:
 |
 |
|
 |