Digital Pathology Lab
Tissue Imaging
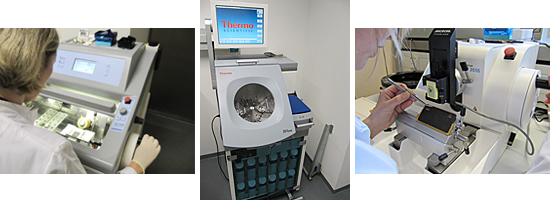
Hamamatsu Nanozoomer Digital Pathology (NDP) System
For automated tissue slide scanning the Hamamatsu NanoZoomer Digital
Pathology (NDP) system is used. It allows the automated scanning of up
to 210 standard slides in one batch with a mean scanning time of
approximately 1 minute per slide (scanning a 15 mm x 15 mm region in
20x magnification) with an additional slide setup time of approximately
1 minute. The specimen can be scanned with either 20x or 40x
magnification yielding a high spatial resolution of 0.46 µm/pixel and
0.23 µm/pixel respectively. This high speed as well as sensitivity is
achieved by using a Time Delay Integration (TDI) high-speed sensor and
scanning the specimen line-by-line. The 3 CCD detector gives superior
colour reproduction of tissue samples being able to discriminate even
between slight colour changes.
In addition to the NDP standard brightfield equipment this
scanner also includes optional fluorescence illumination optics
allowing also the digitisation of fluorescence labelled samples.
After scanning the high resolution-images with a total of approximately
2 GB are presented on a dedicated slide image webserver and can be
viewed comfortably from any workstation using the viewer software from
Hamamatsu. For further analysis e.g. image analysis the images can be
compressed and converted to any desired image format.

- Fast and Automated scanning
- High resolution images (up to 0.23 µm/pixel)
- Fluorescence option
Tissue Analysis
Tissue Analysis Section
Currently, there's a great need for the quantitative characterization of protein expression in human tissues. These efforts, usually denoted antibody proteomics, allow the systematic application of protein-specific antibodies to functionally explore the proteome. Affinity reagents can be utilized in numerous forms, including in vitro and in vivo protein profiling. Affinity reagents can for example be spotted onto microarrays enabling protein arrays. Tissue microarrays allow the high-throughput profiling of specific antigen-antibody interactions. Especially, the computational analysis of large tissue sections allows obtaining comprehensive, statistically much safer statements, than can be obtained from the analysis of limited small microscopic images.
Generally, antibody proteomics is the key component for linking the molecular and the tissue abstraction level. Goal of the TIGA Center is to provide solutions for capturing and describing the spatial expression patterns of proteins in tissues. The coupling of functional systems biological models of protein expression with the computational image analysis of tissues offers new chances for the understanding of key processes of tissue differentiation and tissue homeostasis in general. This approach also offers the chance of practically improving diagnostics and therapeutics in daily routine. Last but not least, the analysis of tissues also is a fundamental prerequisite for a well-grounded computational modeling of tissues.
Histology Lab
Histology Lab inside the TIGA Center
An optimal preparation of histological sections is the basis for their subsequent automatic staining and microscopy. Therefore the TIGA-Center hosts a dedicated histological pipline comprising controlled tissue fixation, standardized sectioning and fully automated staining: Native tissues or organotypic tissue cultures are either cryopreserved or get fixated by using the tissue processor STP-420D capable of fixating 360 samples in one batch. After fixation the tissues are paraffin embedded via the lab own embedding station. In a next step reproducible sections of cryosamples and FFPE-tissues are made by using a microtome or cryotome. For reproducible stainings the TIGA-center runs two Bond-Max staining machines capable to perform fully automated stainings of 60 sections in parallel. Currently the TIGA-center has staining protocols for over 100 antibodies established, ranging from single staining of proliferation and differentiation markers up dedicated double stainings to define and detect tumor infiltrating leukocytes.